Six ways to Sunday: approaches to computational reproducibility in non-model system sequence analysis. презентация
Содержание
- 2. Hello! Assistant Professor; Microbiology; Computer Science; etc. More information at: ged.msu.edu/
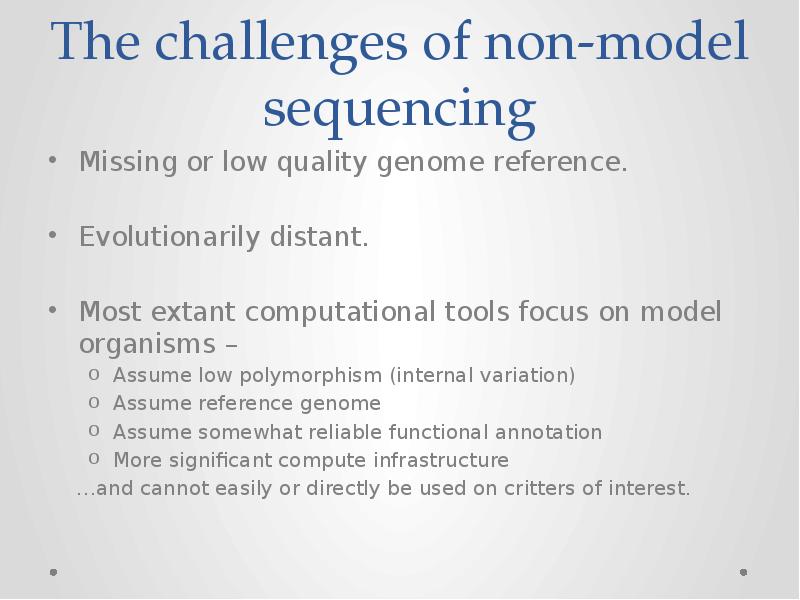
- 3. The challenges of non-model sequencing Missing or low quality genome reference.
- 4. Shotgun sequencing & assembly
- 5. Shotgun sequencing analysis goals: Assembly (what is the text?) Produces new
- 6. Assembly It was the best of times, it was the wor
- 8. Introducing k-mers
- 9. K-mers give you an implicit alignment CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC CATGGACCGATTGCACTGGACCGATGCACGGTACCG
- 10. K-mers give you an implicit alignment CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC CATGGACCGATTGCACTGGACCGATGCACGGTACCG CATGGACCGATTGCACTGGACCGATGCACGGACCG
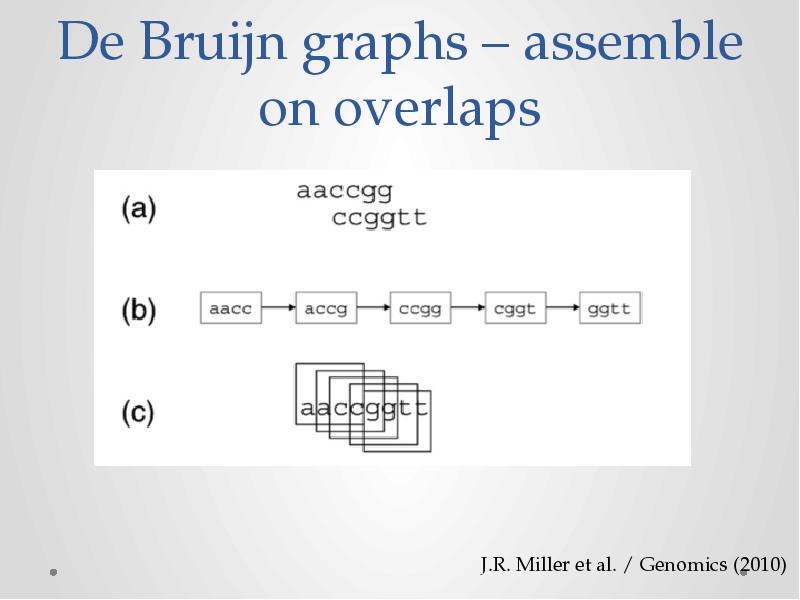
- 11. De Bruijn graphs – assemble on overlaps
- 12. The problem with k-mers CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC CATGGACCGATTGCACTCGACCGATGCACGGTACCG
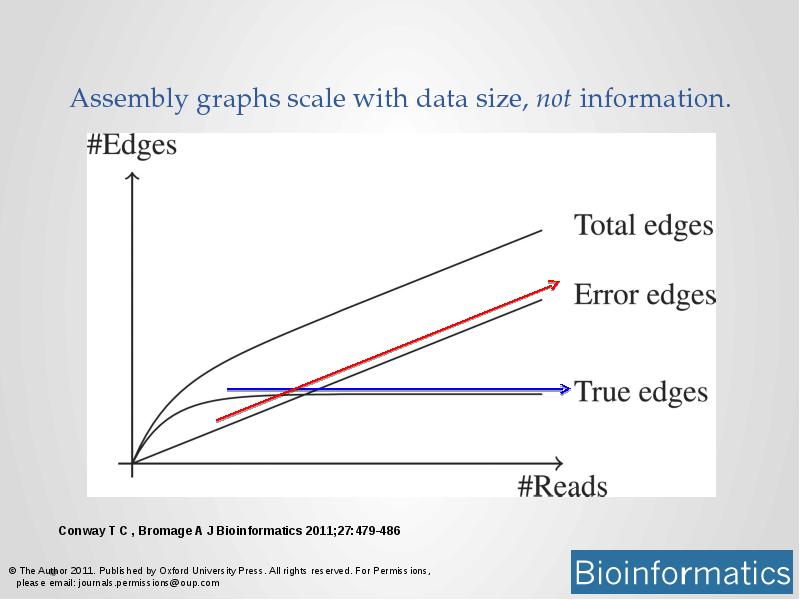
- 13. Assembly graphs scale with data size, not information.
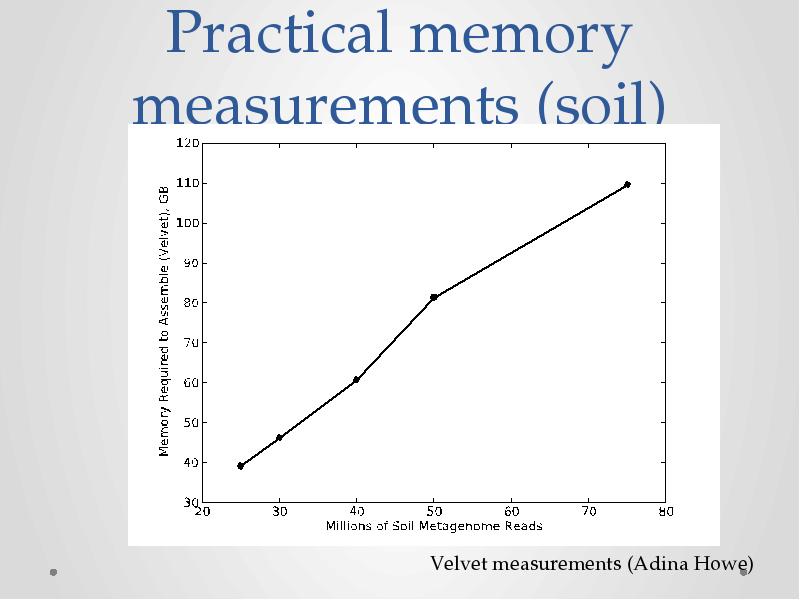
- 14. Practical memory measurements (soil)
- 15. Data set size and cost $1000 gets you ~100m “reads”, or
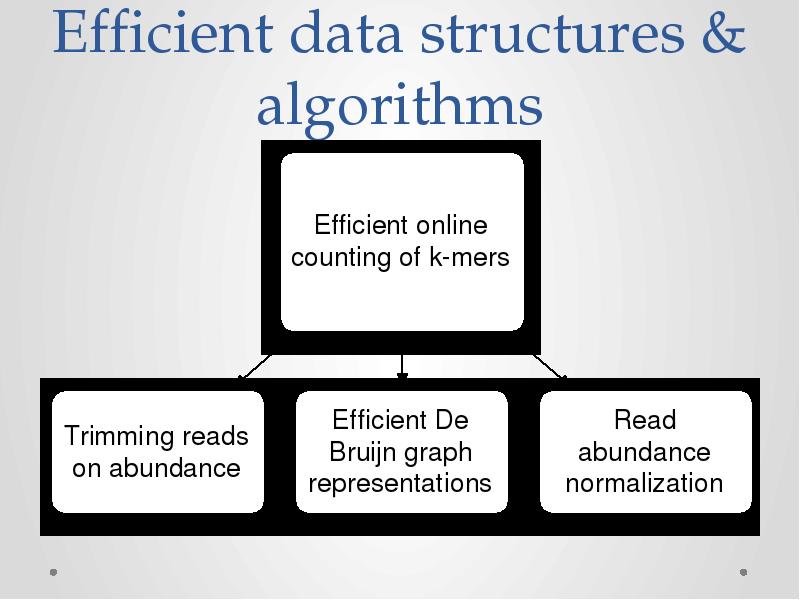
- 16. Efficient data structures & algorithms
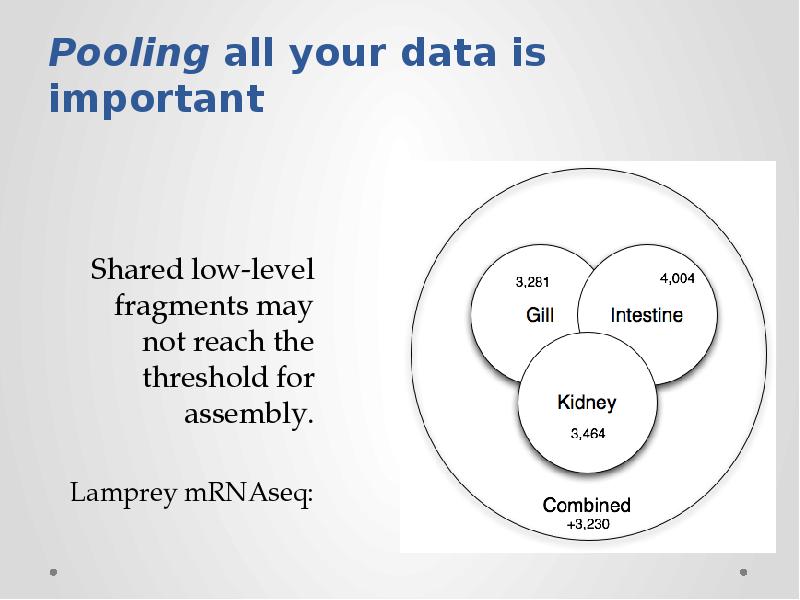
- 17. Shotgun sequencing is massively redundant; can we eliminate redundancy while retaining
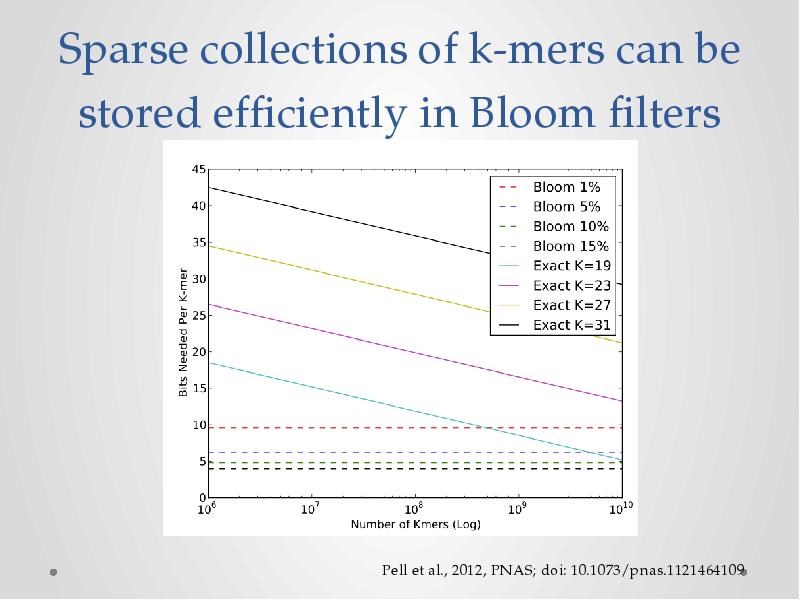
- 18. Sparse collections of k-mers can be stored efficiently in Bloom filters
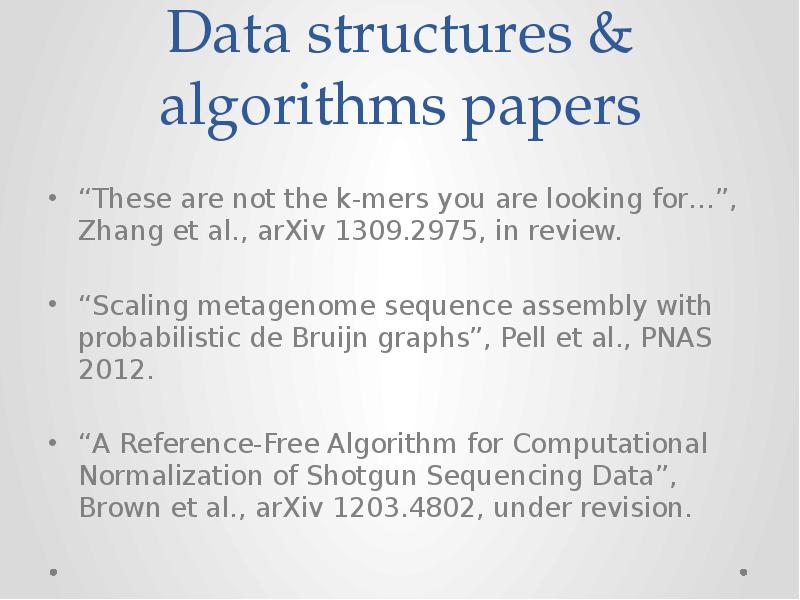
- 19. Data structures & algorithms papers “These are not the k-mers you
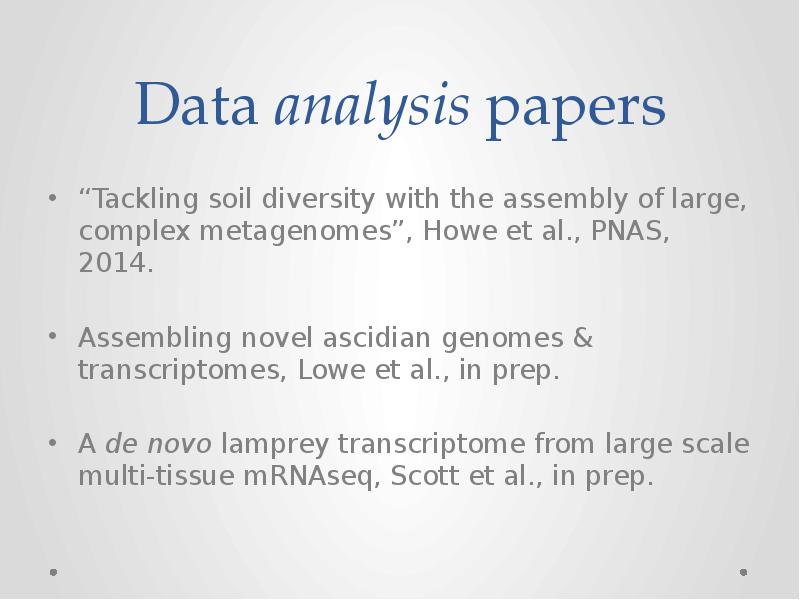
- 20. Data analysis papers “Tackling soil diversity with the assembly of large,
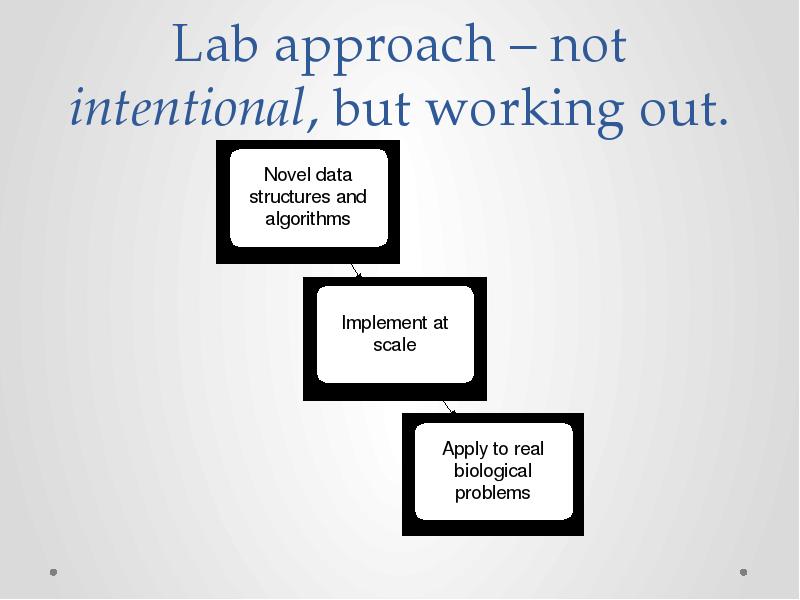
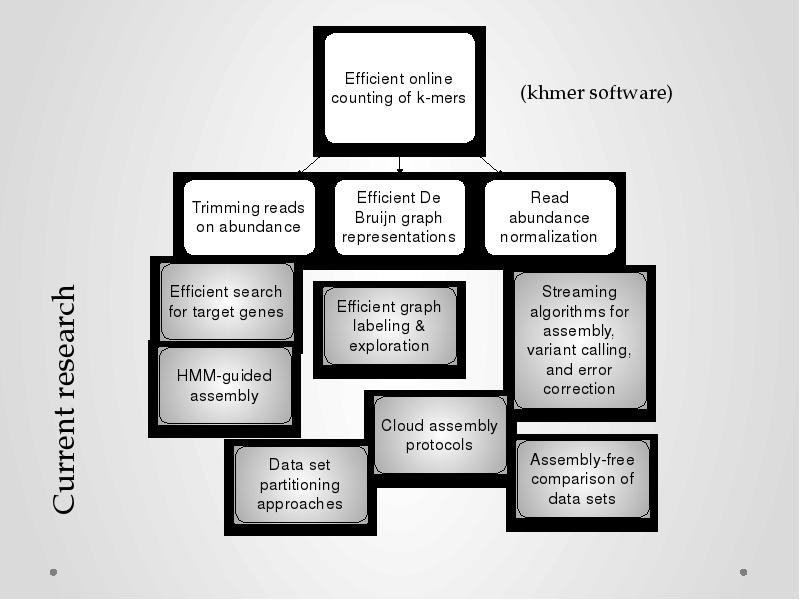
- 21. Lab approach – not intentional, but working out.
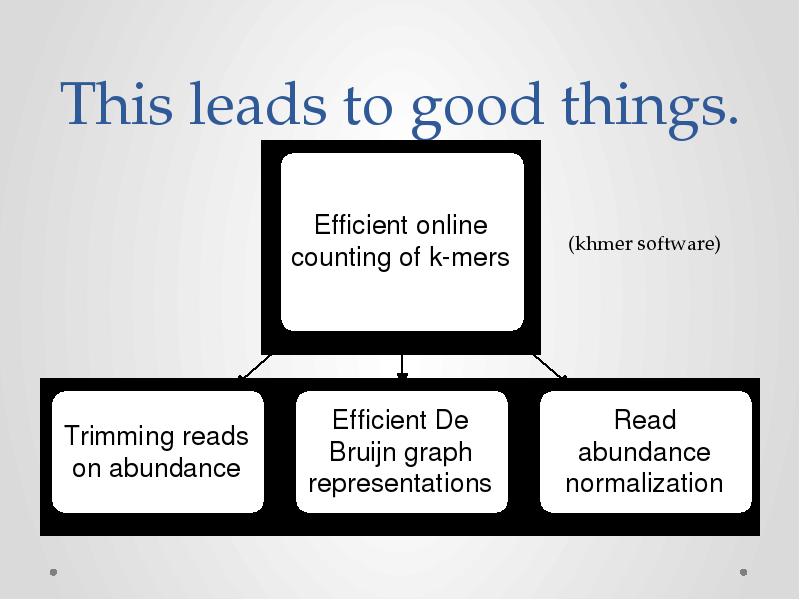
- 22. This leads to good things.
- 24. Testing & version control – the not so secret sauce High
- 25. On the “novel research” side: Novel data structures and algorithms; Permit
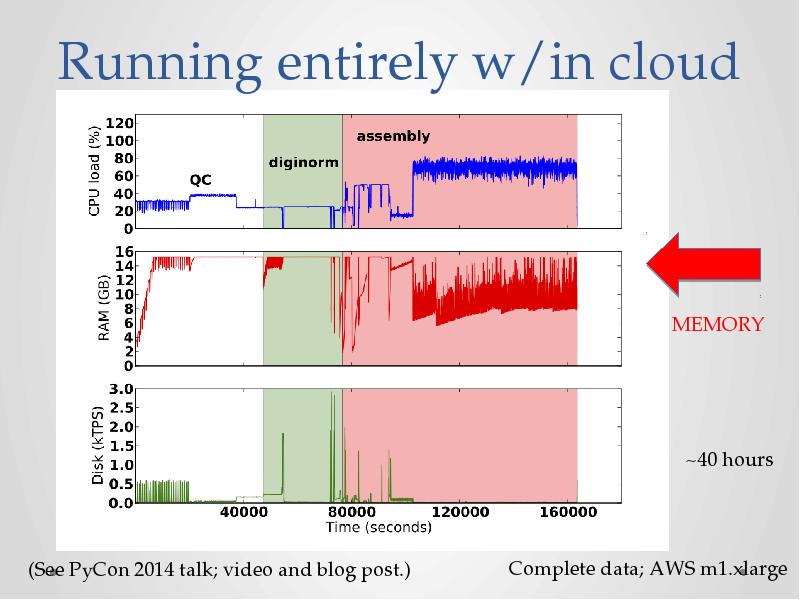
- 26. Running entirely w/in cloud
- 27. On the “novel research” side: Novel data structures and algorithms; Permit
- 28. Reproducibility! Scientific progress relies on reproducibility of analysis. (Aristotle, Nature, 322
- 29. Disclaimer Not a researcher of reproducibility! Merely a practitioner. Please
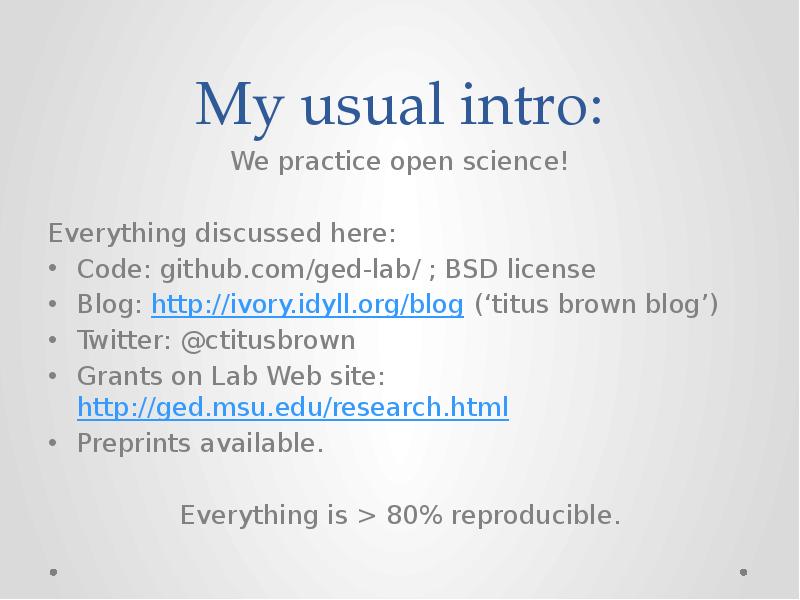
- 30. My usual intro: We practice open science! Everything discussed here: Code:
- 31. My usual intro: We practice open science! Everything discussed here: Code:
- 32. My lab & the diginorm paper. All our code was already
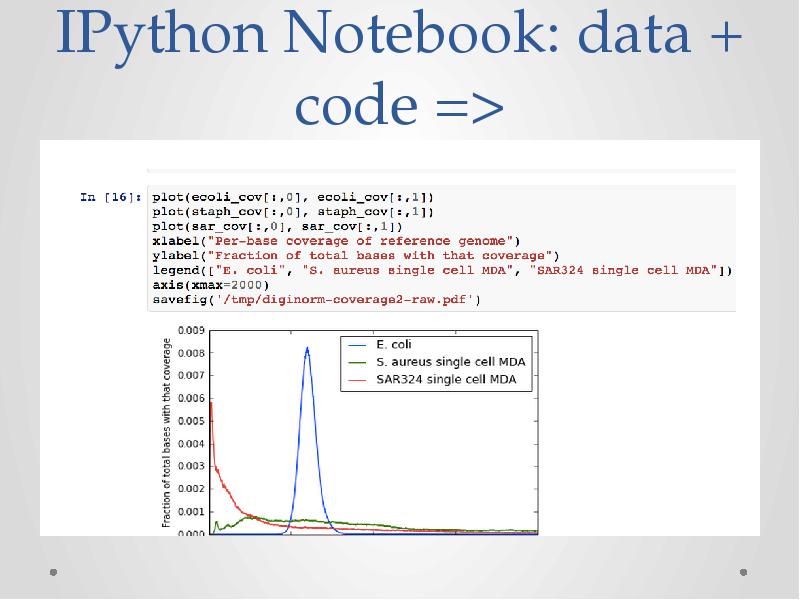
- 33. IPython Notebook: data + code =>
- 34. My lab & the diginorm paper. All our code was already
- 35. To reproduce our paper: git clone <khmer> && python setup.py install
- 36. Now standard in lab --
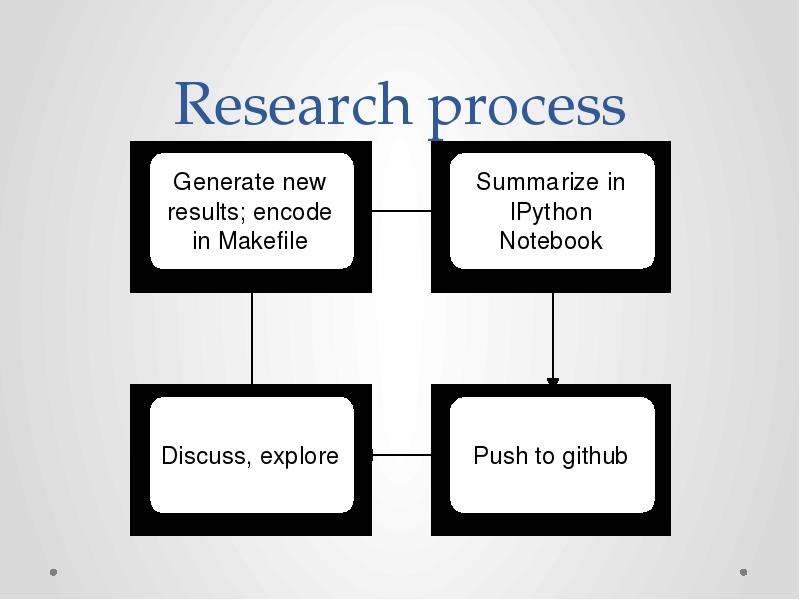
- 37. Research process
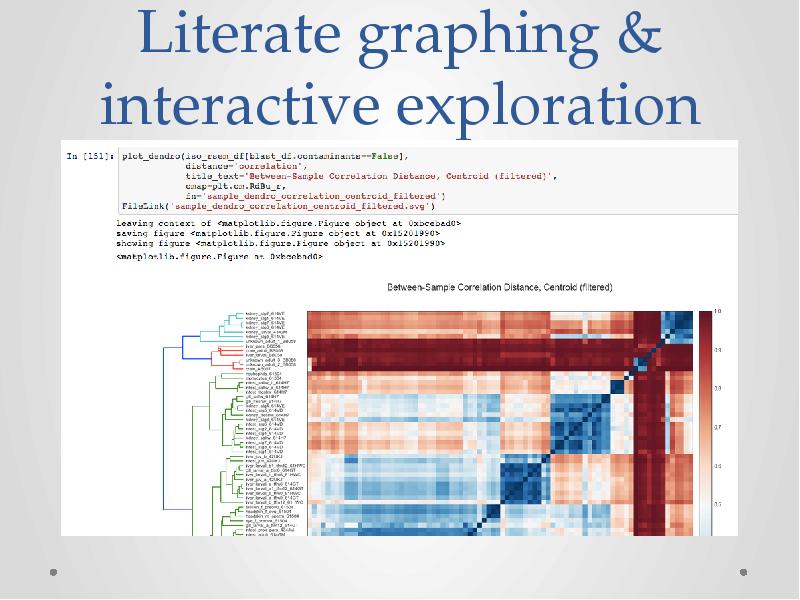
- 38. Literate graphing & interactive exploration
- 39. The process We start with pipeline reproducibility Baked into lab culture;
- 40. Growing & refining the process Now moving to Ubuntu Long-Term Support
- 41. 1. Use standard OS; provide install instructions Providing install, execute for
- 42. 2. Automate Literate graphing now easy with knitr and IPython Notebook.
- 43. Myths of reproducible research (Opinions from personal experience.)
- 44. Myth 1: Partial reproducibility is hard. “Here’s my script.” =>
- 45. Myth 2: Incomplete reproducibility is useless Paraphrase: “We can’t possibly reproduce
- 46. Myth 3: We need new platforms Techies always want to build
- 47. Myth 4. Virtual Machine reproducibility is an end solution. Good start!
- 48. Myth 5: We can use GUIs for reproducible research (OK, this
- 49. Our current efforts? Semantic versioning of our own code: stable command-line
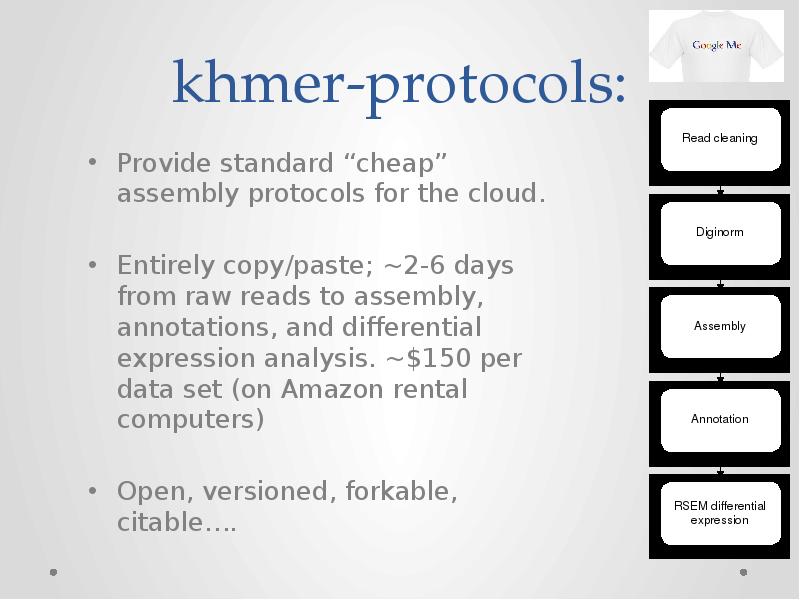
- 50. khmer-protocols
- 51. khmer-protocols: Provide standard “cheap” assembly protocols for the cloud. Entirely copy/paste;
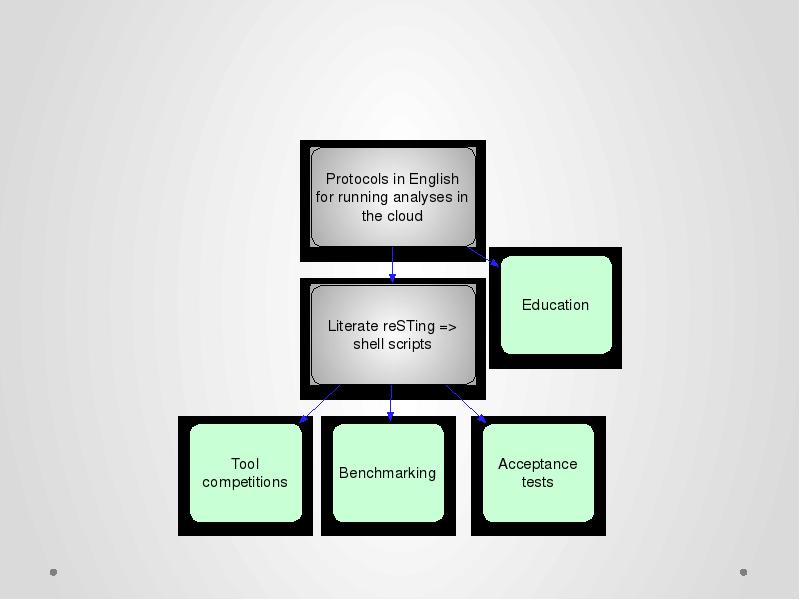
- 52. Literate testing Our shell-command tutorials for bioinformatics can now be executed
- 53. Doing things right => #awesomesauce
- 54. Concluding thoughts We are not doing anything particularly neat on the
- 55. What bits should people adopt? Version control! Literate graphing! Automated “build”
- 56. More concluding thoughts Nobody would care that we were doing things
- 57. Biology & sequence analysis is in a perfect place for reproducibility
- 58. Thanks! Talk is on slideshare: slideshare.net/c.titus.brown E-mail or tweet me: ctb@msu.edu
- 59. Скачать презентацию



































Слайды и текст этой презентации
Скачать презентацию на тему Six ways to Sunday: approaches to computational reproducibility in non-model system sequence analysis. можно ниже:
Похожие презентации